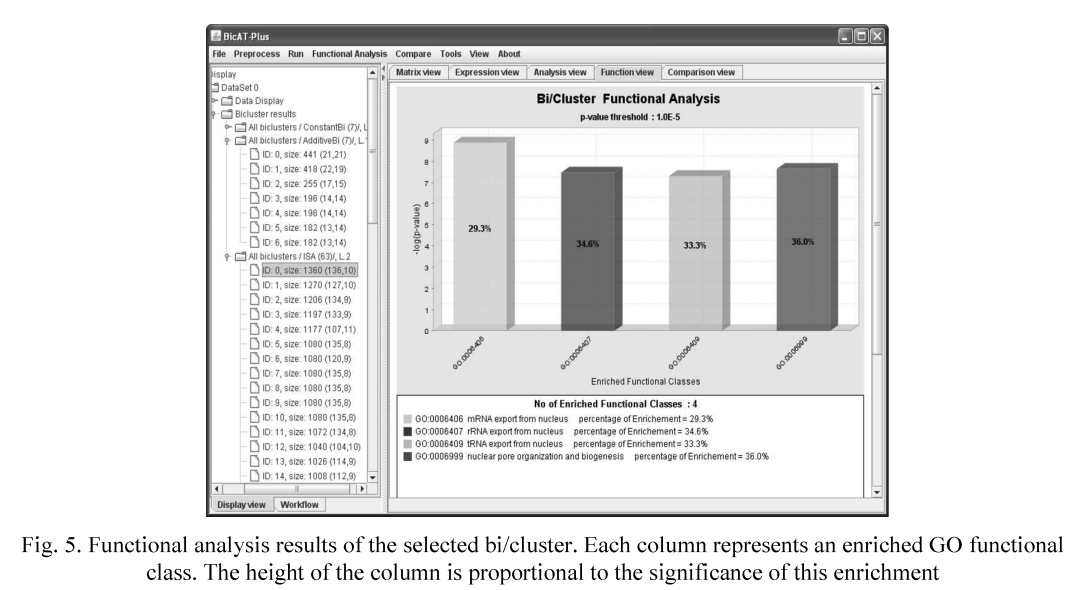
BicATPlus: An automatic comparative tool for Bi/Clustering of gene expression data obtained using microarrays
In the last few years the gene expression microarray technology has become a central tool in the field of functional genomics in which the expression levels of thousands of genes in a biological sample are determined in a single experiment. Several clustering and biclustering methods have been introduced to analyze the gene expression data by identifying the similar patterns and grouping genes into subsets that share biological significance. However, it is not clear how the different methods compare with each other with respect to the biological relevance of the biclusters and clusters as well as with other characteristics such as robustness and predictability. This research describes the development of an automatic comparative tool called BicATplus that was designed to help researchers in evaluating the results of different bi/clustering methods, compare the results against each others and allow viewing the comparison results via convenient graphical displays. BicAT plus incorporates a reasonable biological comparative methodology based on the enrichment of the output bi/clusters with gene ontology functional categories. No exact algorithm can be considered the optimum one. Instead, bi/clustering algorithms can be used as integrated techniques to highlight the most enriched biclusters that help biologists to draw biological prediction about the unknown genes.