Sequencing and assembly of the Egyptian buffalo genome
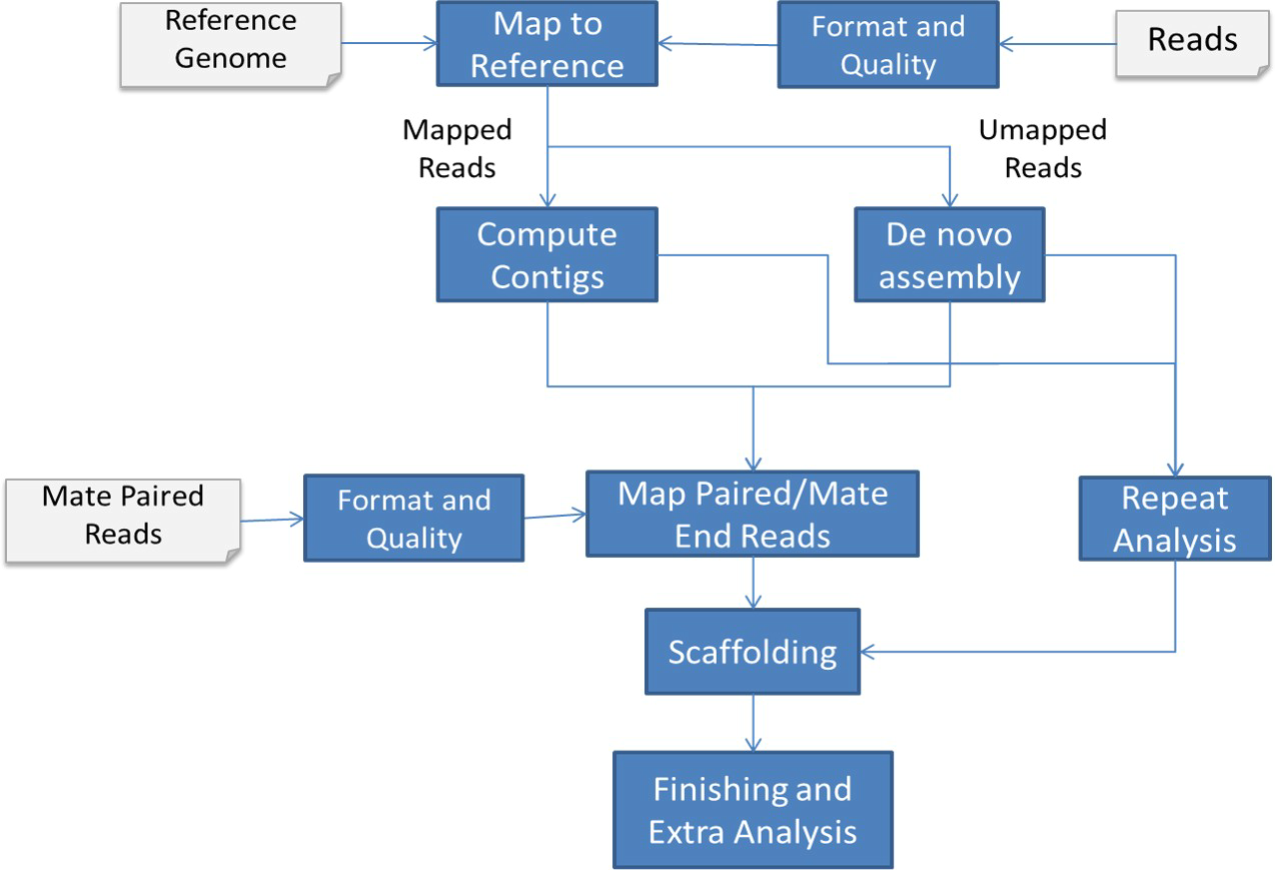
Water buffalo (Bubalus bubalis) is an important source of meat and milk in countries with relatively warm weather. Compared to the cattle genome, a little has been done to reveal its genome structure and genomic traits. This is due to the complications stemming from the large genome size, the complexity of the genome, and the high repetitive content. In this paper, we introduce a high-quality draft assembly of the Egyptian water buffalo genome. The Egyptian breed is used as a dual purpose animal (milk/meat). It is distinguished by its adaptability to the local environment, quality of feed changes, as well as its high resistance to diseases. The genome assembly of the Egyptian water buffalo has been achieved using a reference-based assembly workflow. Our workflow significantly reduced the computational complexity of the assembly process, and improved the assembly quality by integrating different public resources. We also compared our assembly to the currently available draft assemblies of water buffalo breeds. A total of 21,128 genes were identified in the produced assembly. A list of milk virgin-related genes; milk pregnancy-related genes; milk lactation-related genes; milk involution-related genes; and milk mastitis-related genes were identified in the assembly. Our results will significantly contribute to a better understanding of the genetics of the Egyptian water buffalo which will eventually support the ongoing breeding efforts and facilitate the future discovery of genes responsible for complex processes of dairy, meat production and disease resistance among other significant traits. © 2020 El-Khishin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.